L0 segmentation
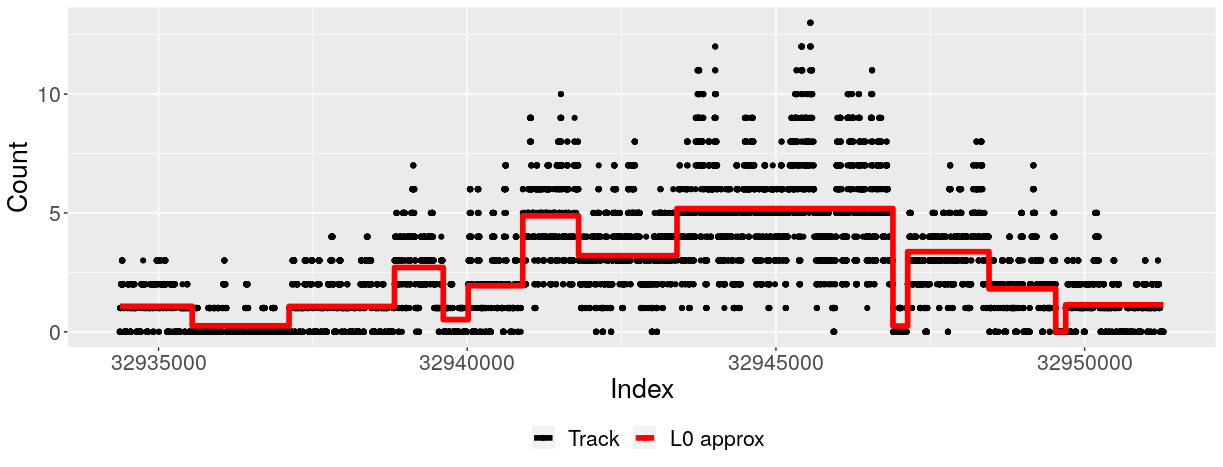
An ultra-fast solution for the the L0 segmentation problem for discovering features from complex epigenetic signals or any sequential data
code.Paper: A unified hypothesis-free feature extraction framework for diverse epigenomic data
PCnt
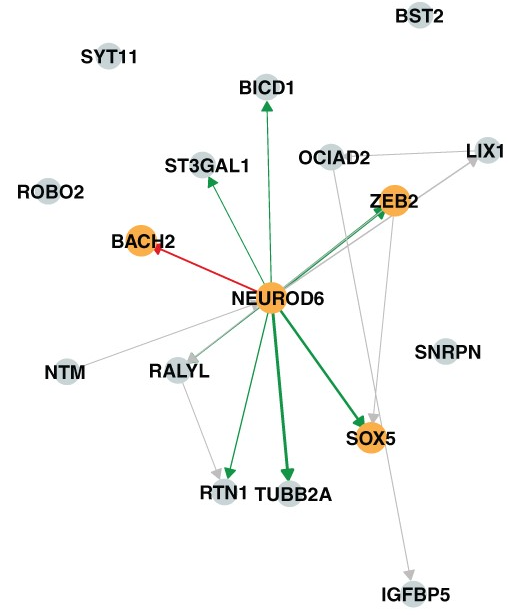
PCnt is a hybrid optimization method for causal learning that combines the strengths of PC and NOTEARS and shows superior performance on real data biological benchmarks
code.InstaPrism

A fast re-implementation of a highly preformant proportion estimation method: BayesPrism
code.Paper: InstaPrism: an R package for fast implementation of BayesPrism
Hetergeneous bulk RNAseq simulation
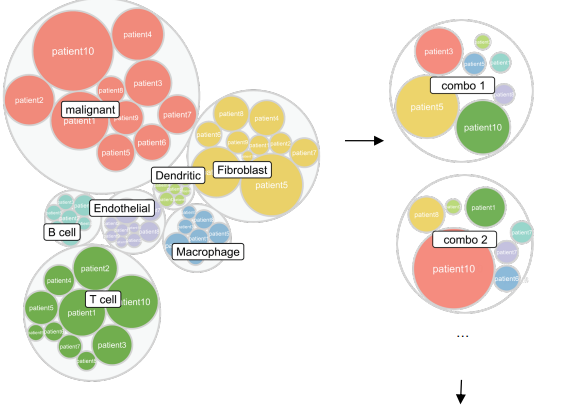
A framework for simulating realistic bulk data from single cell to enable accuarte cell type proportion and deconvolution benchmarking
code.TISFM: totally interpretable sequence to function model
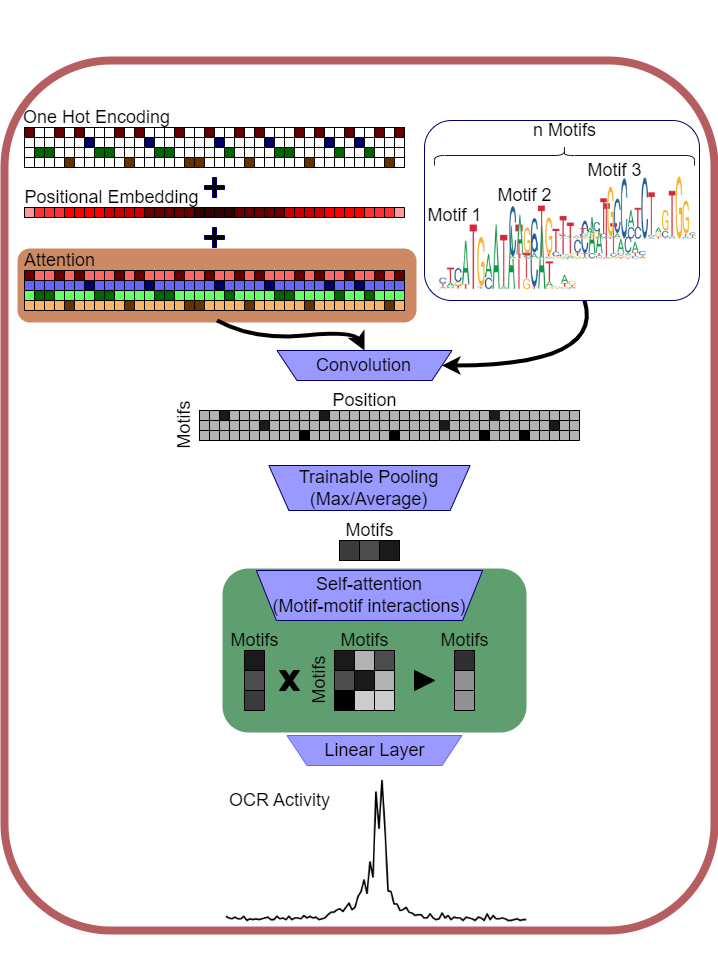
An intrinsically interpretable neural network architecture for sequence-to-function modeling that replaces convolution towers with enitrely interpretable layers and transformations
code.Paper: TISFM: totally interpretable sequence to function model
NIFA Non-negative Independent Factor Analysis
A model that generalizes non-negative matrix factorization (NMF) and independent component analysis (ICA) to find disentangled representations of single cell data
code.
PLIER Pathway-Level Information Extractor
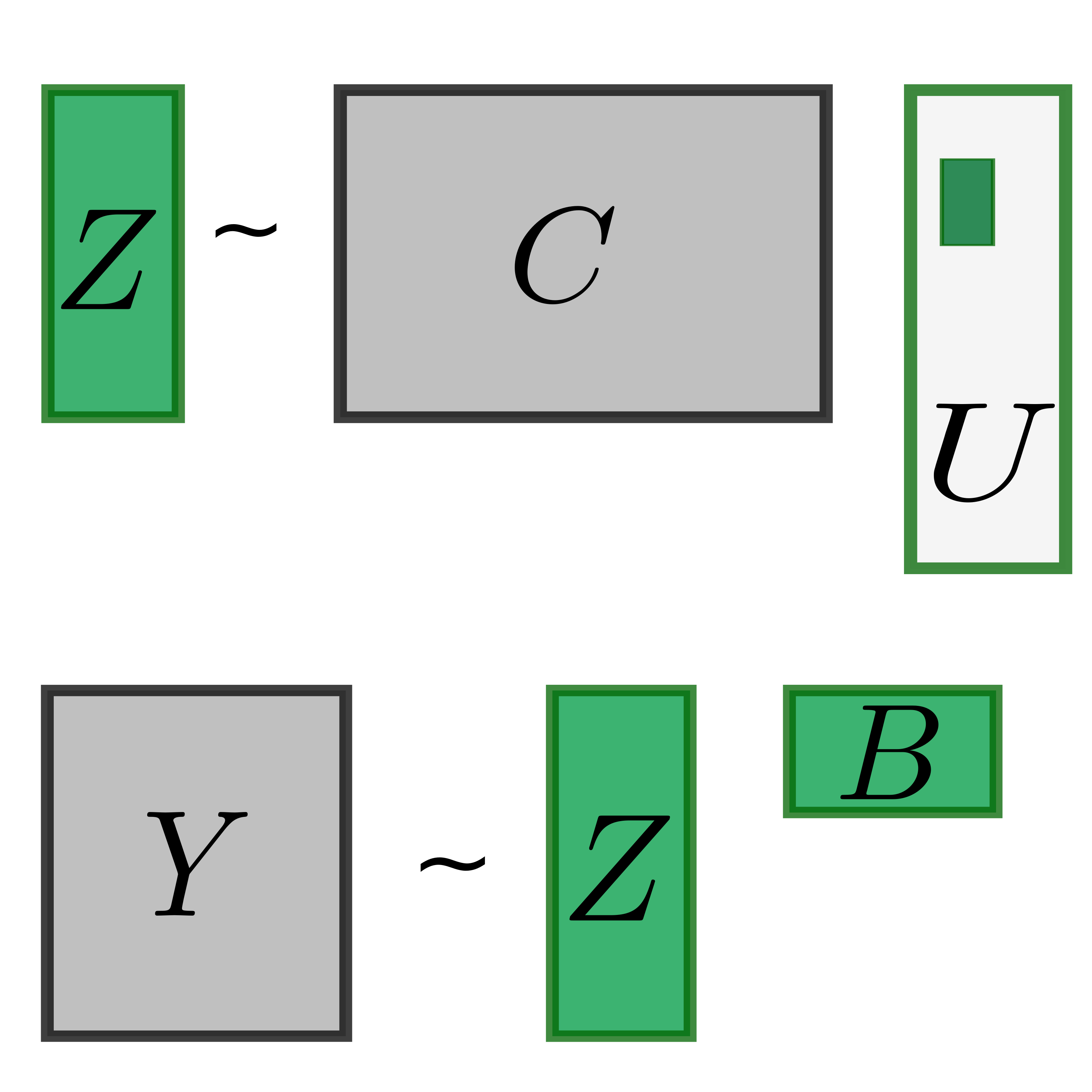
PLIER is a matrix decomposition method that uses prior information from pathway databases to find an interpretable latent variable representation of gene expression datasets.
code.
Paper: Pathway-Level Information ExtractoR (PLIER): a generative model for gene expression data
RERconverge
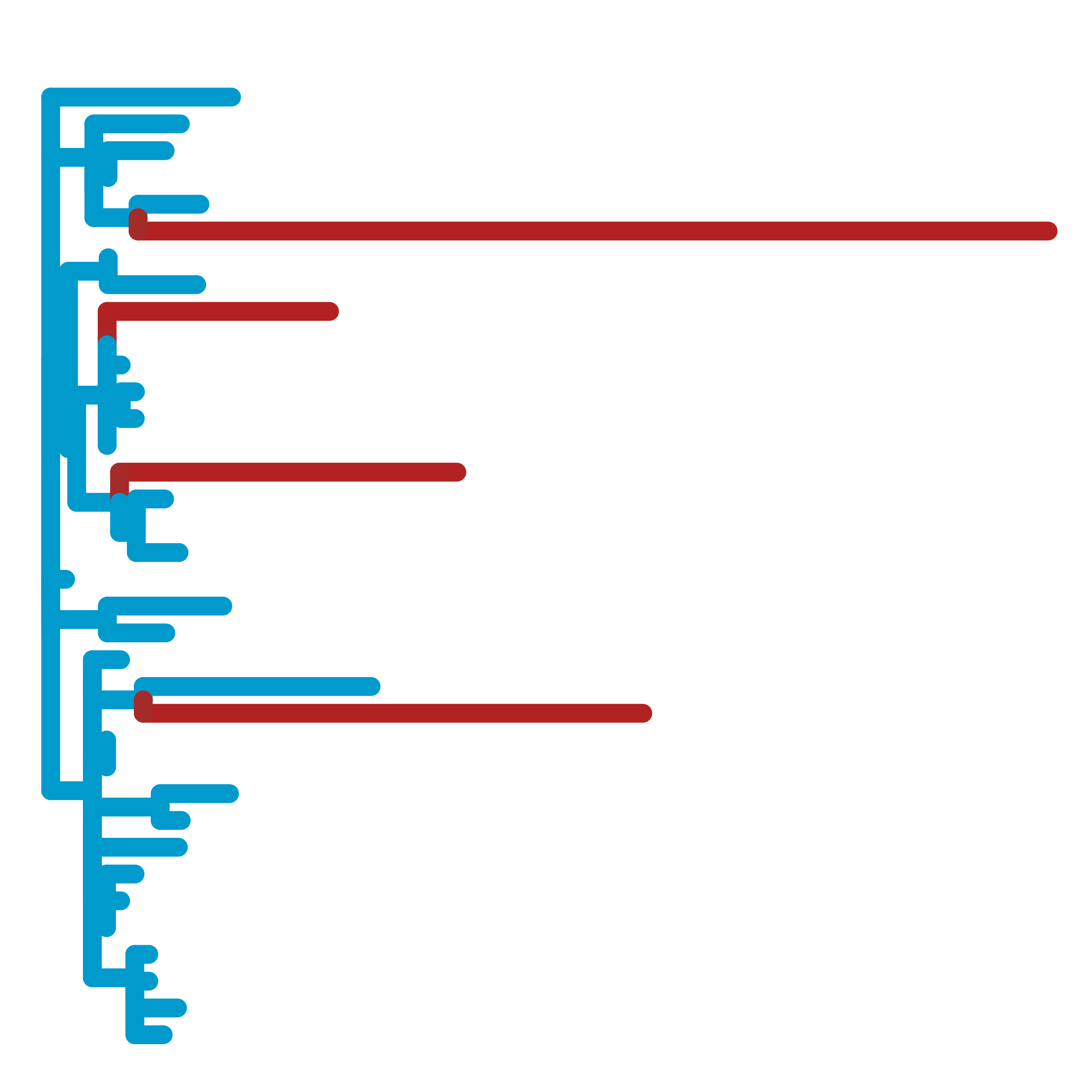
A suite of tools to calculate relative evolutionary rates (RERs) and their associations with phenotypes.
Application papers:
Hundreds of Genes Experienced Convergent Shifts in Selective Pressure in Marine Mammals
Subterranean mammals show convergent regression in ocular genes and enhancers, along with adaptation to tunneling
CELLCode
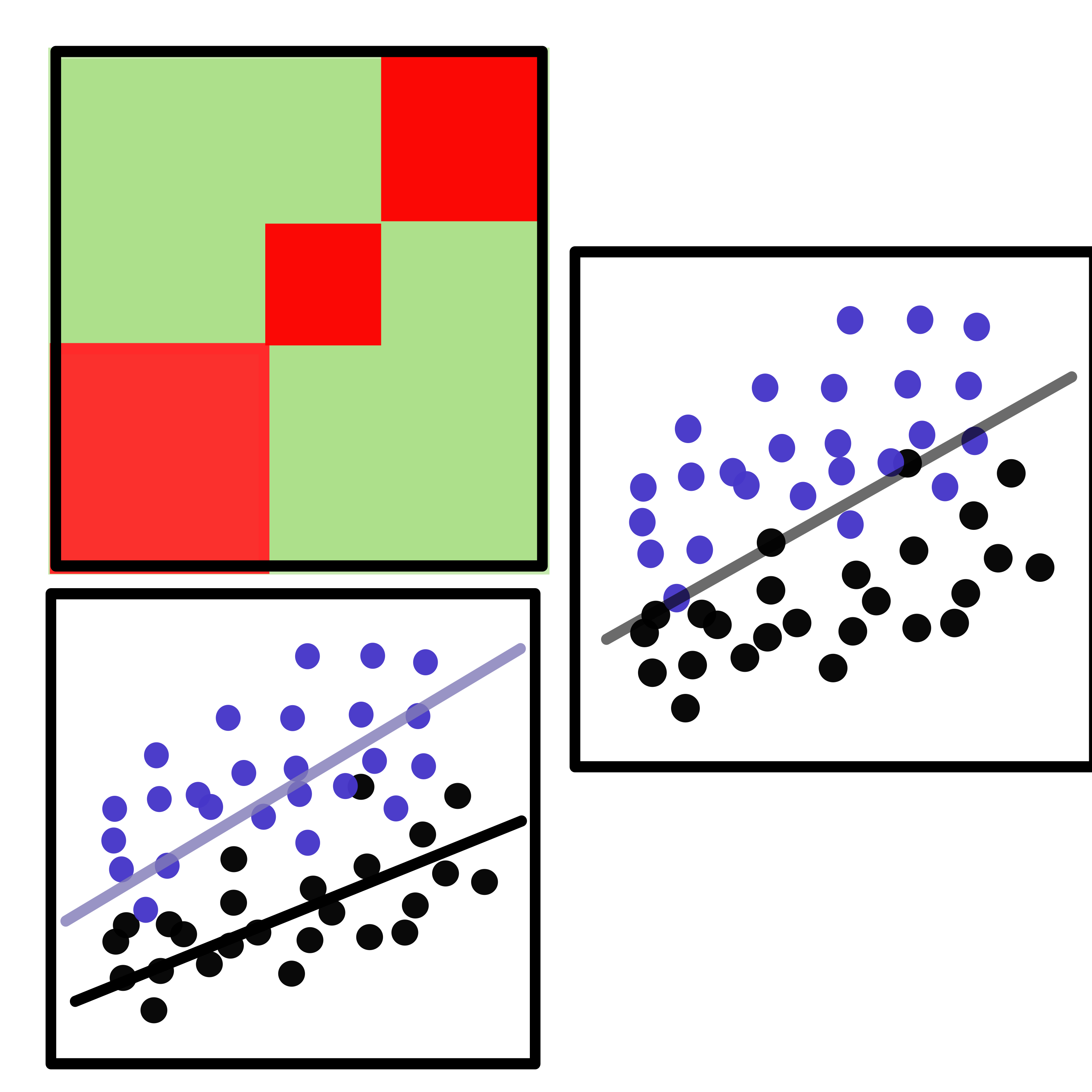
An R package that performs multi-layered differnetial expression analysis to account for tissue composition heterogeneity. It estimates cell-proportions, performs and correction, and assigns trascriptionally regulated genes to the tissue of origin.
DataRemix
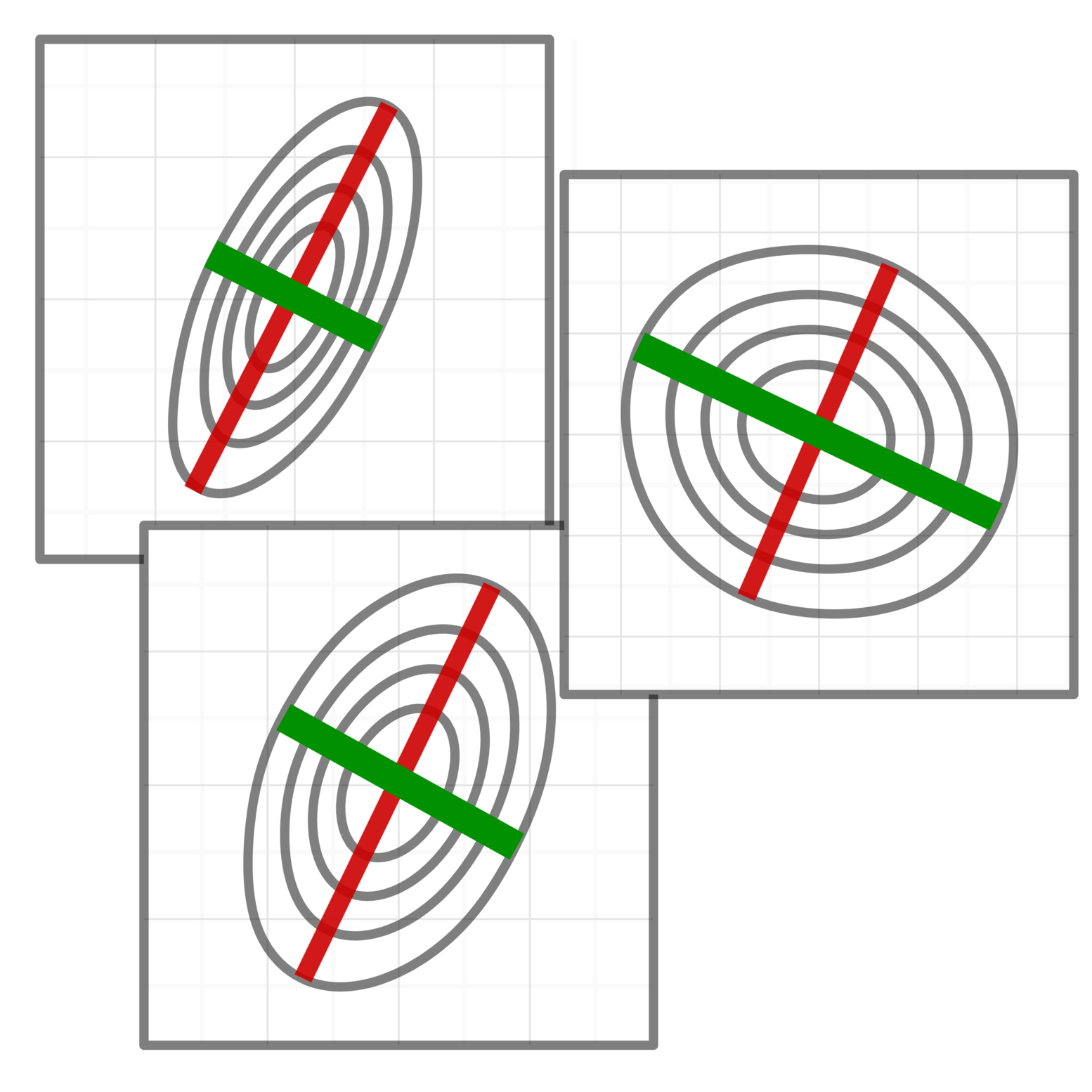
An R package to optimize a data-normalization transform for specific biological tasks.
IntervalStats
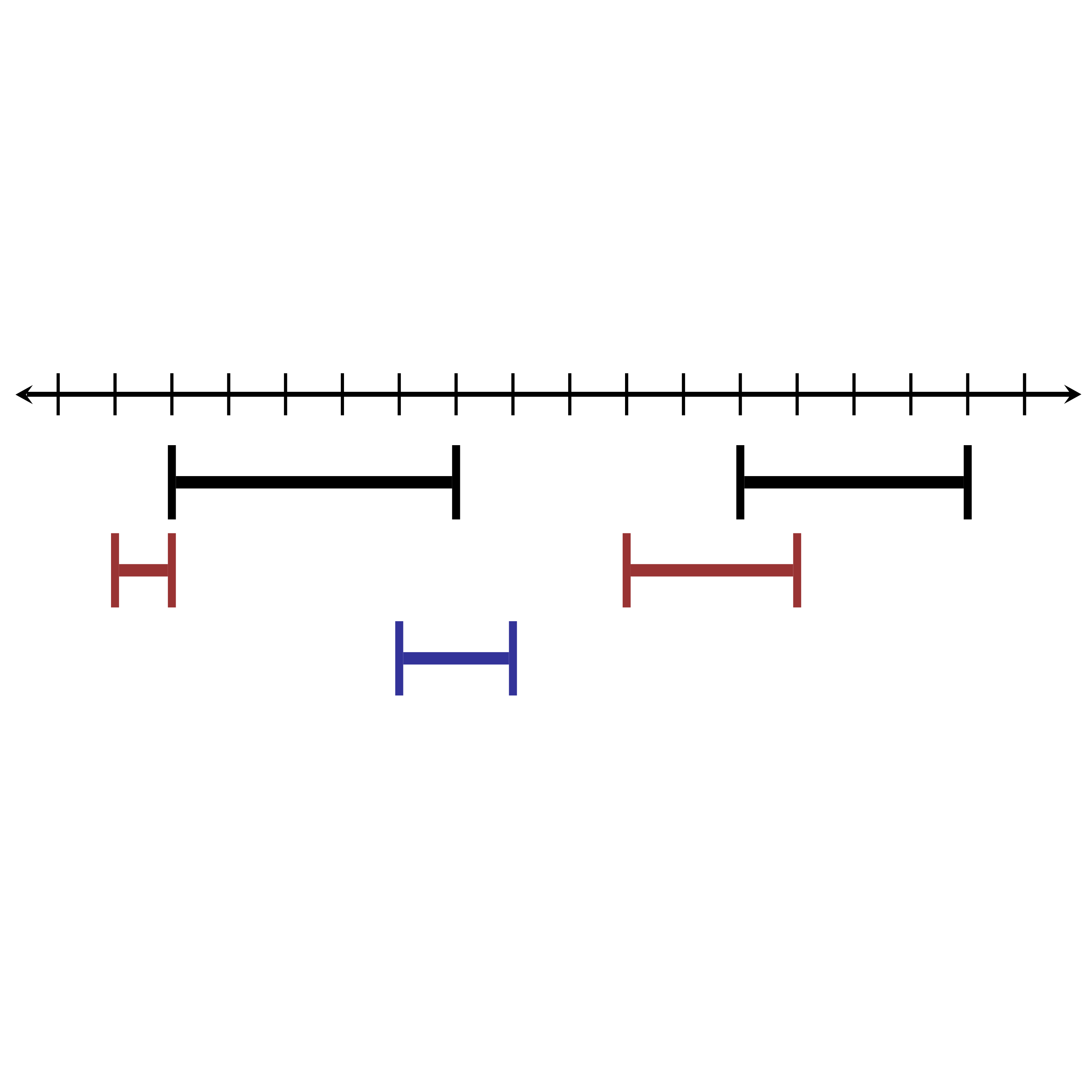
A tool to compute associations between genomic interavals such as peaks for a ChIPseq or ATACseq dataset that uses exact enumeration to compute accurate p-values.
code Also available as part of the coloc-stats webserver.
Paper: An effective statistical evaluation of ChIPseq dataset similarity
EPIANN
An attention-based deep learning model to predict interacting chromosomal regions.
code