Our lab leverages extensive genomic datasets to build machine learning models that advance discovery and deepen interpretative insights across biology. We address a broad spectrum of biological questions, spanning multi-omics analysis to molecular evolution.
Our Home

University of Pittsburgh School of Medicine
Department of Computational and Systems Biology
3501 Fifth Ave,
Pittsburgh, PA 15260
Carnegie Mellon - University of Pittsburgh
Ph.D. Program in Computational Biology

News
- “Causal network perturbations for instance-specific analysis of single cell and disease samples” with Kristina Buschur and Takis Benos is now online at Bioinformatics
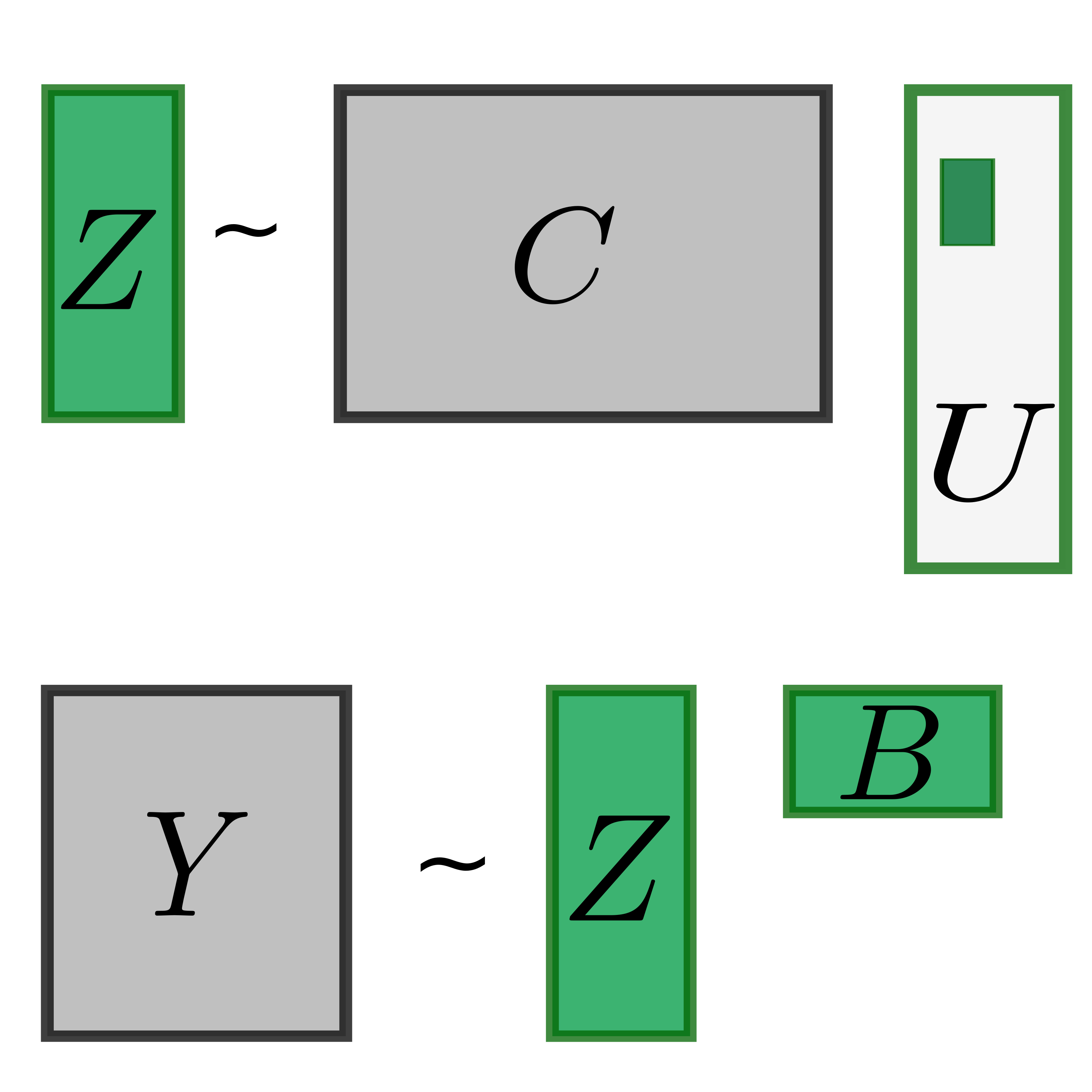
- PLIER is used to infer gene-enironment eQTL interactions. Using Transcriptomic Hidden Variables to Infer Context-Specific Genotype Effects in the Brain
- Our method PLIER is online at Nature Methods. Nature Methods
- Wynn's paper is online at Science With write-ups in NYtimes and National Geographic
Contact Maria
phone: 412 648 3338
email: mchikina@pitt.edu